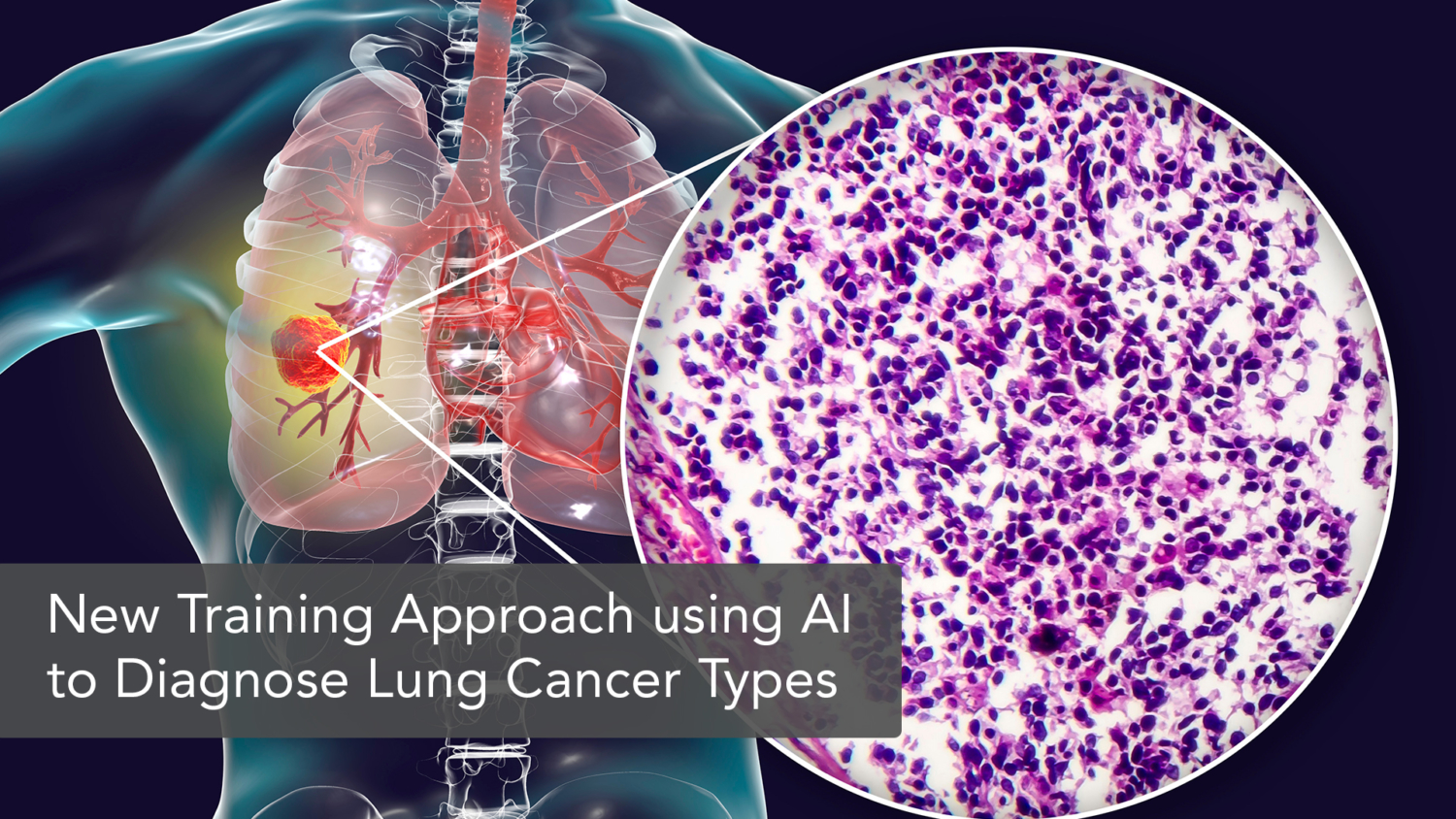
A novel annotation-free whole-slide classification of lung cancer pathologies
Dr. CHEN, Cheng-Yu

Vice President, Taipei Medical University
Dean, College of Humanities and Social Sciences
Director, Research Center of Artificial Intelligence in Medicine and Health
Professor, Department of Radiology
Professor, Professional Master Program in Artificial Intelligence in Medicine, College of Medicine

Summary
The annotation-free whole slide training approach is superior to the multiple-instance learning in classifying non-small cell lung cancer subtypes.
Original Research Article: An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning
Artificial intelligence (AI) is an effort to employ computer science into machines to emulate human thoughts and emotions. In the past decade, AI has achieved incredible growth in an attempt to bridge the gap between human capabilities and machine intelligence. Computer vision is one of the domains in AI that was developed to enable machines to view and perceive the world as a human does. The application extends to image recognition, image analysis, classification, and many more. One particular algorithm that was developed and perfected was the convolutional neural network (CNN). It is a deep learning algorithm that represents the input of an image, extracts the importance of every bit of the image (each neuron), and distinguishes one image from another. In the medical field, deep learning algorithms have shown performances matching human capacity in various diagnostic imaging tasks that assists in disease identification, including cancer.
In nonsmall-cell lung cancer (NSCLC), proper pathological diagnosis to differentiate between adenocarcinoma and squamous cell carcinoma presents a challenge, as the difference in pathological features between them is subtle. CNN has been the primary method used in image recognition in distinguishing these two histologically different NSCLC subtypes. Despite its success in tumor type identification and classification, the patch-based CNN method demands substantial annotations on the slides by experienced pathologists. Multiple-instance learning (MIL), another previous technique employed to reduce the annotation burden, on the other hand, is subject to incorrect selection of patches for diagnosis or classification in early training leading to compromise in the model’s performance. To overcome the limitations of existing methods, a group of researchers from Taipei Medical University (TMU) proposed the use of whole slide imaging (WSI) as means to train CNN utilizing slide-level labels. Based on the outcome, the WSI method is proven to be superior to the existing methods of digital diagnosis, specifically in comparison to the MIL method.
For the purpose of establishing an optimum approach for pathological classification of lung cancer subtypes, these researchers developed a annotation-free whole slide training method to train CNN. The design consolidates unified memory (UM) mechanism and some graphics processing unit (GPU) optimization technique to train standard CNN with much bigger image input without compromising training pipeline or model architecture. A total of 7003 slides were gathered, consisting of 2039 cases of non-cancer, 3876 of adenocarcinoma and 1088 cases of squamous carcinoma, then randomly assigned for training, validation, and testing purposes. Subsequently, the slides were tested using multiple performance models to study different parameters in image analysis in lung cancer. As a result, the WSI method used recorded superior results to MIL in visualization, small lesion diagnosis, image resolution (despite data size), and memory consumption. Such superiority is attributed to two things, the randomness of the sampling and the existence of ceiling to the performance of MIL due to the lack of ground truth at patch level. Therefore, a heftier sample size might be required in future research in comparing the two different models. Provided that there is also a limitation on host memory when training larger images on CNN, future research on a more memory-efficient algorithm will further optimize the model in digital diagnosis of diseases.
First introduced in 1999, WSI has achieved remarkable growth in technology and is recognized in various applications in pathological diagnosis ever since. The approval of the US FDA to a WSI system for primary surgical pathology diagnosis in 2017 has paved the way for the acceptance of this technology for broader medical applications. The application of AI in pathology is most often limited by the constrain of annotation that retards the process pace due to its focus at a microscopic level. The method explored in this study can counter that disadvantage by providing rapid progression via reduction of meticulous annotation. Given that the healthcare demand is increasing by the day, non-laborious, highly efficient technology is snatching importance in enhancing diagnosis and disease classification processes. Thus, the utilization of WSI to train CNN in the diagnosis and classification of lung cancer becomes imminently crucial and contributes to the realization of United Nations Sustainable Development Goal (UNSDG) 3, good health and well-being. Given the promising future advancement of WSI technology and numerous benefits, WSI should be adopted and prioritized for its application in the healthcare sector by authority bodies and investors. Considering that cost is a current limitation for its implementation, a more cost-effective WSI should be developed to facilitate the medical fraternity to contribute to the overall healthcare improvement of the global community.
1. Kumar N, Gupta R, Gupta S. Whole Slide Imaging (WSI) in Pathology: Current Perspectives and Future Directions. Journal of Digital Imaging. 2020;33(4):1034-40.
2. Saha S. A Comprehensive Guide to Convolutional Neural Networks — the ELI5 way. 2018.
For interviews or a copy of the paper, contact Office of Global Engagement via global.initiatives@tmu.edu.tw.