Sorting Genomes with Software:
Maxbin’s Development and Future
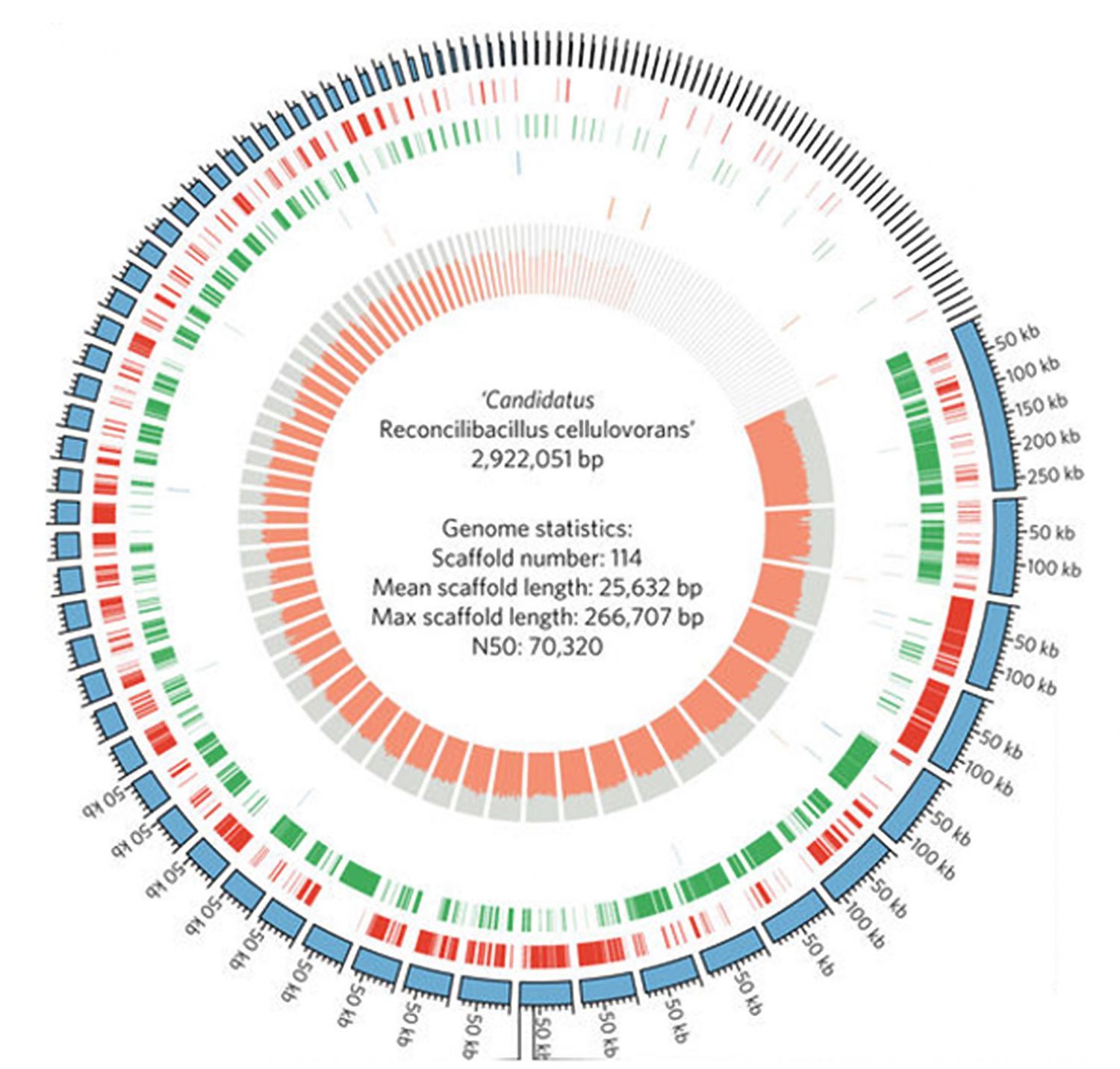
The traditional process of studying the microbiome (the aggregation of single celled organisms in a certain niche, like compost, your gut, or even the cup of coffee you’re drinking) is slow and labor intensive. More importantly, 95 to 99% of organisms can’t be successfully cultured in this way.
Dr. Yu-Wei Wu, an assistant professor at TMU’s Graduate Institute of Biomedical Informatics, has taken a different approach in metagenomics – the study of the microbiome – with his Maxbin software. “[We] take all the DNA from everything at the same time, then we try to analyze their diversity and how they constitute, collaborate, or compete with each other. It’s really like playing a jigsaw puzzle, because you cannot really tell where the DNA is coming from.” Maxbin is an automated algorithm that sorts genomic sequences from metagenomic datasets – a process known as binning.
The idea for Maxbin struck Dr. Wu during a shower in 2013. “I’m a computational biologist, and I’m a lazy one. So I don’t like to do these manual things and I want to try to make everything automatic. So I came up with an idea to integrate machine learning into this process.” The next day he shared his idea with his supervisor, Dr. Steve Singer at the Earth Sciences division of the Lawrence Berkeley National Laboratory (Berkeley Lab). “I wasn’t sure if he really understood my idea, but he said just go ahead”
A month later a prototype was ready, and a year later he had a workable tool for sorting and identifying the genomic material present in microbiomes. The original version focused on examining one sample at a time and didn’t address cross sample comparisons. After constant developments, the 2.0 version can now integrate multiple samples of up to 1,000 organisms and produce a highly accurate genome. The algorithm is unsupervised, needing no prior database information or other assistance to perform classification.
Maxbin’s first use was binning DNA from samples of plant based compost to develop of better biofuels. The team at Berkley tested several hundred organisms to see which were able to best tolerate the toxic environments involved fuel production, and came up with at least six promising organisms. Maxbin continues to be used in ongoing experiments at the U.S. Department of Energy’s (DOE) Joint BioEnergy Institute (JBEI).
On the medical front, researchers are beginning to probe the relationships between the human microbiome and disease. Promising avenues of medical research involves finding bacteria types from DNA, genetic functions from RNA, and protein compounds produced by bacteria that can be correlated with patient recovery in patients with gut diseases such as diabetes and obesity, or correlated in a clinical setting with medical imaging data.
At Taipei Medical University’s Microbiota Core Facility, human microbiota samples are being collected for use by medical researchers. Dr. Wu has already been approached by physicians at TMU’s affiliated hospitals to examine microbiome correlations with lung disease, and has had success using Maxbin and artificial intelligence to predict antibiotic resistance.
At present Maxbin needs some manual checks to confirm data quality, but Dr. Wu is working to increase accuracy using machine learning and artificial intelligence to the point where manual checks are unnecessary. Together with upgrades to computing hardware, these improvements will reduce processing times that for complex samples with many microorganisms might take several days.
Further software refinements will allow Maxbin to account for organisms that make up a smaller fraction of samples, and for the algorithm to be applied to other forms of biological information. “We need to see all 4 aspects [DNA, RNA, proteins, and compounds] to tell a good story about who the microbes are and what are they really doing in the environment, and what they are producing, and how do they compete [or collaborate] with each other.”
Thanks to Dr. Wu and Maxbin, scientists have a tool that significantly reduces the complexity of microbiome analysis.
Dr. Wu is committed to keeping Maxbin available to the public for use in biomedical research. Maxbin 2.0 is available for download at www.sourceforge.net/projects/maxbin2/.
For interviews or a copy of the paper, contact Office of Global Engagement via global.initiatives@tmu.edu.tw.